For nearly a century, evolutionary biology has been shaped by Modern Synthesis, or Neo-Darwinism. This framework rests on the idea that changes in DNA sequences, mutations, are the fundamental drivers of adaptation and the vast diversity of life we see today. Through natural selection and genetic drift, genetic variation is filtered, refined, and passed down across generations.
But a new study published in Nature Plants suggests that the story of evolution has another chapter. Researchers have found that plants can influence traits and evolve without altering their DNA sequence at all. Instead, they use chemical modifications called methylation tags. Much like sticky notes on the pages of a book, these tags do not rewrite the text itself but signal which parts are most important to read.
The study was led by Dr. Zaigham Shahzad, Associate Professor at the Department of Life Sciences, SBA School of Science and Engineering, LUMS, and Dr. Daniel Zilberman, Professor at the Institute of Science and Technology, Austria, in collaboration with researchers from John Innes Center, Norwich, UK, and Michigan State University, US. The team studied Arabidopsis thaliana, a small mustard-like plant widely used in biology, to distinguish the effects of genetic mutations from those of epimutations, changes in DNA methylation patterns, on natural phenotypic variation. Analyzing gene expression across more than 10,000 genes in 625 natural genotypes, the researchers showed that heritable DNA methylation explains as much phenotypic diversity as conventional genetic mutations.
Through further genetic analyses, they uncovered that gene body methylation plays a key role in regulating gene expression. By tracking methylation patterns, they could identify genes linked to important traits such as nutrient uptake, flowering time, and plant fitness. Remarkably, their “epigenome-wide association” mapping approach proved 1.5 times more precise than traditional genome-wide association methods.
One striking advantage of this epigenetic system lies in its speed. While genetic mutations occur slowly, epimutations happen at rates 100,000 times higher, allowing plants to experiment with traits without making permanent genetic changes. This ability is particularly valuable in rapidly changing environments.
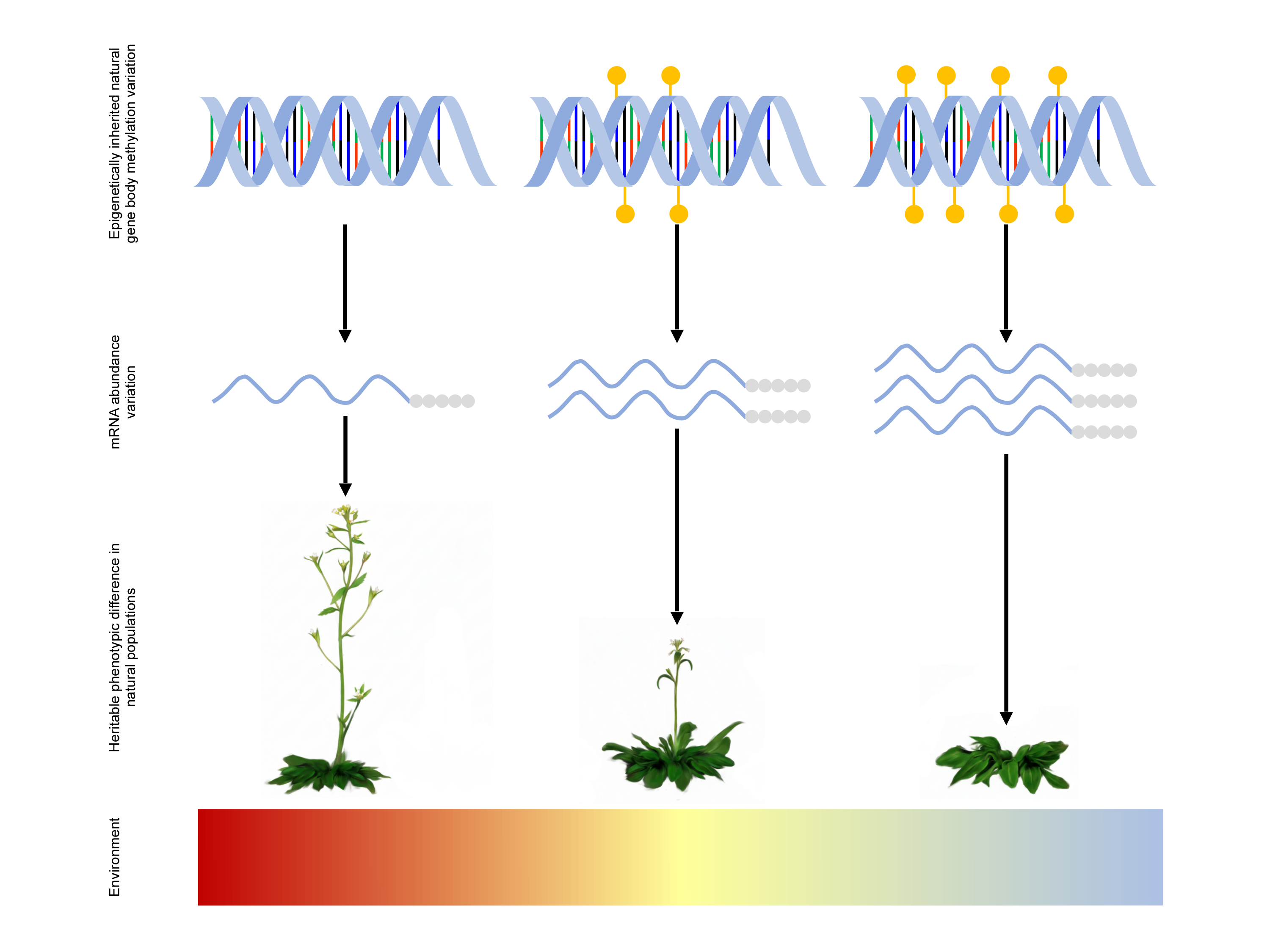
The researchers also found evidence that plants are already putting this system to use. In regions with higher levels of nitrogen dioxide, which is a pollutant released by human activity, Arabidopsis plants flower earlier. This shift, they discovered, is linked to methylation changes at FLC, a well-known flowering control gene. Since the rise of nitrogen dioxide pollution has occurred only over the past few decades, traditional genetic evolution alone would not have kept pace. Epigenetic changes, however, seem fast enough to help plants adapt in real time.
These findings expand our understanding of how life evolves. Beyond DNA sequence changes, organisms have access to a parallel system, one based on chemical tags layered onto existing genes. This research underscores how scientists at SBASSE, LUMS, are collaborating internationally to address fundamental questions in biology, showing that life’s adaptability runs deeper than DNA alone.